
Fractions: inspect from which fractions proteins and peptides are likely to come from.Spectrum IDs: compare the search engine performance and see how the search engine results are combined.Overview: get a simple yet detailed overview of all the proteins, peptides and PSMs in your dataset.PeptideShaker currently supports nine different analysis tasks: It can be used on local data as well as on data sets deposited to the ProteomeXchange PRIDE repository.
PeptideShaker can be used in command line and comes with rich visualization capabilities to navigate the results. PeptideShaker aggregates the results in a single identification set, annotates spectra, computes a consensus score, maps sequences and performs protein inference, scores post-translational modification localization, runs statistical validation, quality control, and annotates the results using multiple sources of information like Gene Ontology, UniProt and Ensembl annotation, and protein structures.

PeptideShaker is a search engine independent platform for interpretation of proteomics identification results from multiple search and de novo engines, currently supporting X!Tandem, MS-GF+, MS Amanda, OMSSA, MyriMatch, Comet, Tide, Mascot, Andromeda, MetaMorpheus, Sage, Novor, DirecTag and mzIdentML.
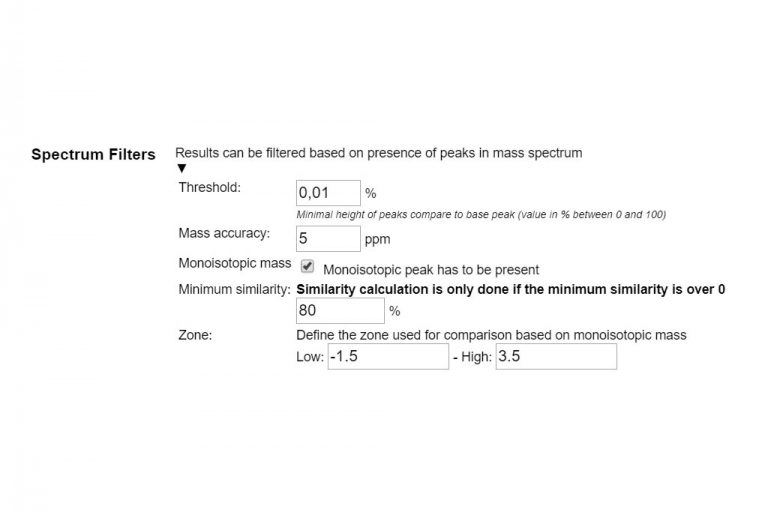
#PEPTIDESHAKER SPECTRAL LIBRARY FULL SIZE#
(Click on an image to see the full size version) If you use PeptideShaker as part of a publication please include this reference.


 0 kommentar(er)
0 kommentar(er)
